TSS assembly pipeline for Dm_EPDnew_001
Introduction
This document provides a technical description of the transcription start site assembly pipeline that was used to generate EPDnew version 001 for Drosophila melanogaster genome assembly dm3.
Source Data
| Description | URLs |
| ENSEMBL64 | Source URL: http://sep2011.archive.ensembl.org
MGA doc: /mga/dm3/ensembl64/ensembl64.html MGA data: /ftp/mga/dm3/ensembl64/dm3_ENSEMBL64.sga.gz |
| MachiBase | Source URLhttp://machibase.gi.k.u-tokyo.ac.jp/
MGA doc: /mga/dm3/machibase/machibase.html MGA data: /ftp/mga/dm3/machibase/all_oligocap.sga.gz |
| CAGE data from Hoskins et a. 2012. PMID: 21177961 | Source URL: http://genome.cshlp.org/content/21/2/182/suppl/DC1 MGA doc: /mga/dm3/hoskins11/hoskins11.html MGA data: /ftp/mga/dm3/hoskins11/embryo_cage.sga.gz |
| H3K4me3 in S2 cells from Gan et al. 2010. PMID: 20398323 | Source URL:
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM480156
MGA doc: /mga/dm3/gan10/gan10.html MGA data: /ftp/mga/dm3/gan10/GSM480156_dm3-S2-H3K4me3.sga.gz |
Assembly pipeline overview
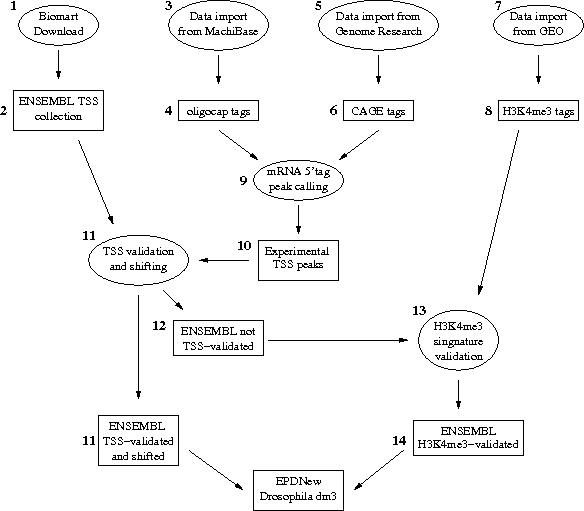
|
Description of procedures and intermediate data files
1. Biomart Download
Data was downloaded from sep2011.archive.ensembl.org/biomart/martview/ selecting the following attributes:- Ensembl Gene ID
- Ensembl Transcript ID
- Chromosome Name
- Strand
- Transcript Start (bp)
- Transcript End (bp)
- Gene Start (bp)
- Gene End (bp)
- Status (transcript)
- Status (gene)
- Associated Gene Name
- Transcripts of protein coding genes only
- Transcript length > 0 [Transcript Start different from Transcript End]
- Transcript lies on full chromosomes
- Gene must have a 5' UTR [Transcript Start different from Gene Start]
- Genes must be annotated [Associated Gene Name present]
- Gene and transcripts status known
Transcripts with the same TSS position were merged under a common ID. As a conseguence of this, from the 23850 transcrips originally present in the ENSEMBL database, 5953 were merged, leaving 17897 uniquely mapped promoters in the input list.
2. EMBL TSS collection
The ENSEMBL TSS collection is stored as a tab-deliminated text file conforming to the SGA format under the name:-
filename.
- NCBI/RefSeq chromosome id
- "TSS"
- position
- strand ("+" or "-")
- "1"
- gene name.
3. Data import from MachiBase
MachiBase data were generated with the oligo-capping technology. The source data were downloaded from: According to the readme file included in the tar archive, the 5' end tags were mapped to the Drosophila genome using BLAT as alignment tool allowing for up to three mismatches.4. oligocap tags
The compressed version of this file is available from the MGA archive (see above) under the name:-
all_oligocap.sga.gz.
5. Data import from Genome Research
Mapped sequence tags were extracted from Supplementary Data File 1 available from Genome Research at:The downloaded source file is in SAM format and has been generated with the tag mapping program StatMap as described in the article cyted above. We extracted all tags with mapping quality scores greater or equal to 30.
6. CAGE tags
The compressed version of this file is available from our ftp site (see above link) with the name:-
embryo_cage.sga.gz
7. Data import from GEO
BED files for the GEO serie GSE19325 were downloaded from GEO ftp site and converted into SGA file using in house software.8. H3K4me3 tags
The compressed version of this file is available from our ftp site (see above link) with the name:-
GSM480156_dm3-S2-H3K4me3.sga.gz
9. mRNA 5' tags peak calling
CAGE tags and oligocap tags have been merged into a unique file with the following command:-
sort -m -s -k1,1 -k3,3n -k4,4 embryo_cage.sga all_oligocap.sga > all_data.sga
- Window width = 100
- Vicinity range = 200
- Peak refine = Y
- Count cutoff = 9999999
- Threshold = 8
11. TSS validation and shifting
The source data (mRNA peaks and ENSEMBL TSS) was then processed in a pipeline aiming at validating transcription start sites with mRNA peaks. An ENSEMBL TSS was experimentally confirmed if an mRNA peak lied in a window of 100 bp around it. The validated TSS was then shifted to the nearest base with the higher tag density. After this step, the total number of validated promoters was 10389.The list of validated and shifted promoters can be downloaded here.
12. ENSEMBL not-validated TSS
The total number of non mRNA validated TSS was 7508. These promoters were not discarded but were the subject of the following validation step.13. H3K4me3 signature validation
H3K4me3 histone mark was used as a marker for promoter validation. The 7508 promoters that were not validated by mRNA signature were scored according to the presence of H3K4me3 histon mark near the TSS. This procedure recover more than 2000 promoters bringing the total number of validated promoters to 12435.15. EPDnew collection
The 12435 experimentally validated promoter were stored in the EPDnew database that can be downloaded from our ftp site. Scientist are wellcome to use our other tools ChIP-Seq (for correlation analysis) and SSA (for motifs analysis around promoters) to analyse EPDnew database.Last update October 2019